Once the molecule file is fully loaded, the image at right will become live. At that time the "activate 3-D" icon ![]() will disappear.
will disappear.
Phenol (C6H5OH)
The
geometry optimizations for the three
highest levels of theory are shown below for phenol. The levels of
theory used were 621G, 631G, and DZV. The buttons below display the bond
lengths and the bond angles for each level of theory respectively. Table 1: This table displays the bond lengths for phenol taken from the literature4.
| Bond Type |
Bond Length in Angstroms |
| C-C |
1.398 |
| C-H othro |
1.084 |
| C-H meta |
1.076 |
| C-H para |
1.082 |
| C-O |
1.364 |
| O-H |
0.956 |
The overall percent error in the total bond lengths was 5.74%.
The vibrational frequencies were calculated using the highest level of theory, DZV. The IR-spectrum for Phenol is displayed below retrieved from the NIST website4.
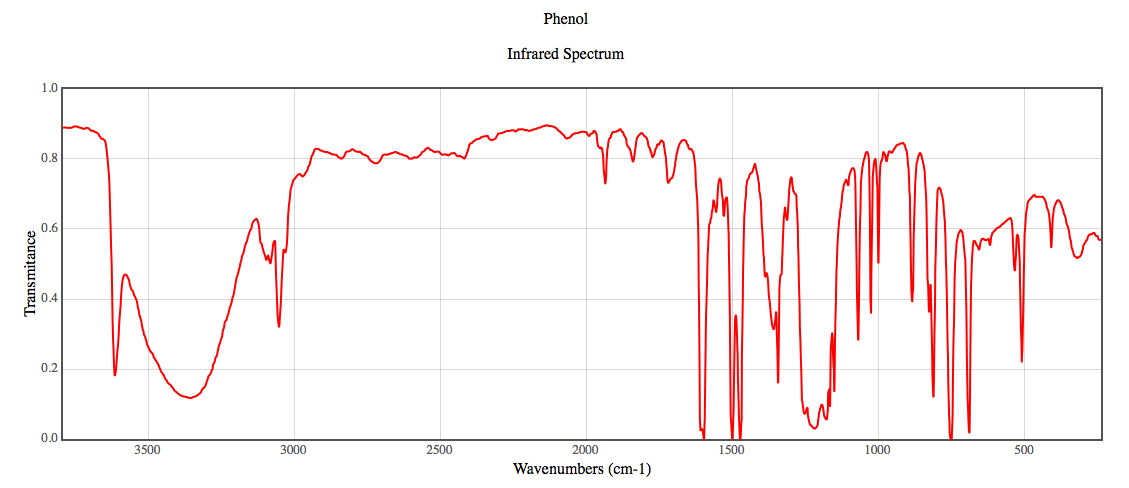
Figure 1:
This is an IR-Spectrum of phenol displaying the vibrational frequencies of the bonds4.
Table 2: This tables displays the vibrational frequencies for the bonds in phenol taken from the literature1.
This is an IR-Spectrum of phenol displaying the vibrational frequencies of the bonds4.
Table 2: This tables displays the vibrational frequencies for the bonds in phenol taken from the literature1.
| Type of Vibration |
Wavelength in cm^-1 |
| C-H bend |
680-900 |
| C=C stretch |
1440-1620 |
| C-H stretch |
3000-3100 |
| O-H stretch |
3350-3650 |
C-H bend
C=C stretch
C-H stretch
O-H stretch
The buttons above display the vibrations calculated from the highest level of theory. All values calculated using DZV were overestimates giving large percent errors of 6.53%, 0.087% (this was a very good estimate), 9.68%, and 11.96% respectively.
UV-Vis spectra values were calculated with the highest level of geometry optimization theory.
Table 3: This table contains values of constants taken from NIST used for UV-Vis calculations below6.
The best optimization for the dipole moment of phenol was calculated from 621G giving a percent error of 17.5%.
The buttons above display the vibrations calculated from the highest level of theory. All values calculated using DZV were overestimates giving large percent errors of 6.53%, 0.087% (this was a very good estimate), 9.68%, and 11.96% respectively.
UV-Vis spectra values were calculated with the highest level of geometry optimization theory.
Table 3: This table contains values of constants taken from NIST used for UV-Vis calculations below6.
| Constant |
Value of Constant |
| Planck's Constant (h) |
6.62606957x10^-34 Js |
| Speed of Light (C) |
2.99792x10^8 m/s |
Energy = Planck's constant x speed of light / wavelength
Wavelength = Planck's constant x speed of light / Energy
Table 6: This table displays the dipole moments of all levels of theory for phenol.Wavelength = Planck's constant x speed of light / Energy
Table 4: This table displays the oscillator strength and wavelengths for phenol.
| Oscillator Strength (unitless) |
Wavelength in nanometers (nm) |
| ground to 1st = 0.046723 |
1979 |
| ground to 3rd = 1.311125 |
1465 |
| ground to 4th = 1.106149 |
1445 |
| 1st to 15th = 0.000002 |
2569 |
The dipole moments of phenol were calculated with all levels of theory and compared to the literature values.
Table 5: This table displays the dipole moment of phenol taken from the literature5.
Table 5: This table displays the dipole moment of phenol taken from the literature5.
| Molecule |
Dipole moment value from literature |
| Phenol |
1.40 +/- 0.03 |
| Level of Geometry Optimization Theory |
Dipole moment value |
| 621G |
1.645541 |
| 631G |
1.767432 |
| DZV |
1.814355 |
The best optimization for the dipole moment of phenol was calculated from 621G giving a percent error of 17.5%.
Page skeleton and JavaScript generated by export to web function using Jmol 14.2.12_2015.01.22 2015-01-22 21:48 on Mar 7, 2015.
This will be the viewer

If your browser/OS combination is Java capable, you will get snappier performance if you use Java